Note
-
Download Jupyter notebook:
https://docs.doubleml.org/stable/examples/py_double_ml_gate.ipynb.
Python: Group Average Treatment Effects (GATEs) for IRM models#
In this simple example, we illustrate how the DoubleML package can be used to estimate group average treatment effects in the DoubleMLIRM model.
[1]:
import numpy as np
import pandas as pd
import doubleml as dml
from doubleml.irm.datasets import make_heterogeneous_data
Data#
We define a data generating process to create synthetic data to compare the estimates to the true effect. The data generating process is based on the Monte Carlo simulation from Oprescu et al. (2019).
The documentation of the data generating process can be found here. In this example the true effect depends only the first covariate \(X_0\) and takes the following form
[2]:
np.random.seed(42)
data_dict = make_heterogeneous_data(
n_obs=500,
p=10,
support_size=5,
n_x=1,
binary_treatment=True,
)
data = data_dict['data']
print(data.head())
y d X_0 X_1 X_2 X_3 X_4 X_5 \
0 6.114530 1.0 0.925248 0.180575 0.567945 0.915488 0.033946 0.697420
1 5.580922 1.0 0.474214 0.862043 0.844549 0.319100 0.828915 0.037008
2 1.278434 0.0 0.696289 0.339875 0.724767 0.065356 0.315290 0.539491
3 1.794805 0.0 0.615863 0.232959 0.024401 0.870099 0.021269 0.874702
4 6.178169 1.0 0.350712 0.767188 0.401931 0.479876 0.627505 0.873677
X_6 X_7 X_8 X_9
0 0.297349 0.924396 0.971058 0.944266
1 0.596270 0.230009 0.120567 0.076953
2 0.790723 0.318753 0.625891 0.885978
3 0.528937 0.939068 0.798783 0.997934
4 0.984083 0.768273 0.417767 0.421357
The generated dictionary also contains the true individual effects saved in the key effects.
[3]:
ite = data_dict['effects']
print(ite[:5])
[4.770944 5.4235839 5.07202564 5.30917769 4.97441062]
The goal is to estimate the average treatment effect for different groups based on the covariate \(X_0\). The groups can be specified as DataFrame with boolean columns. We consider the following three groups
[4]:
groups = pd.DataFrame(
np.column_stack((data['X_0'] <= 0.3,
(data['X_0'] > 0.3) & (data['X_0'] <= 0.7),
data['X_0'] > 0.7)),
columns=['Group 1', 'Group 2', 'Group 3'])
print(groups.head())
Group 1 Group 2 Group 3
0 False False True
1 False True False
2 False True False
3 False True False
4 False True False
The true effects (still including sampling uncertainty) are given by
[5]:
true_effects = [ite[groups[group]].mean() for group in groups.columns]
print(true_effects)
[np.float64(2.906716732639898), np.float64(5.223485956098176), np.float64(4.827938162750831)]
Interactive Regression Model (IRM)#
The first step is to fit a DoubleML IRM Model to the data.
[6]:
data_dml_base = dml.DoubleMLData(
data,
y_col='y',
d_cols='d'
)
[7]:
# First stage estimation
from sklearn.ensemble import RandomForestClassifier, RandomForestRegressor
ml_g = RandomForestRegressor(n_estimators=500)
ml_m = RandomForestClassifier(n_estimators=500)
np.random.seed(42)
dml_irm = dml.DoubleMLIRM(data_dml_base,
ml_g=ml_g,
ml_m=ml_m,
n_folds=5)
print("Training IRM Model")
dml_irm.fit()
print(dml_irm.summary)
Training IRM Model
coef std err t P>|t| 2.5 % 97.5 %
d 4.482349 0.086826 51.624206 0.0 4.312172 4.652526
Group Average Treatment Effects (GATEs)#
To calculate GATEs just call the gate() method and supply the DataFrame with the group definitions and the level (with default of 0.95). Remark that for straightforward interpretation of the GATEs the groups should be mutually exclusive.
[8]:
gate = dml_irm.gate(groups=groups)
print(gate.confint(level=0.95))
2.5 % effect 97.5 %
Group 1 2.701491 3.016702 3.331913
Group 2 5.097140 5.314781 5.532421
Group 3 4.412864 4.669562 4.926260
The confidence intervals above are point-wise, but by setting the option joint and providing a number of bootstrap repetitions n_rep_boot.
[9]:
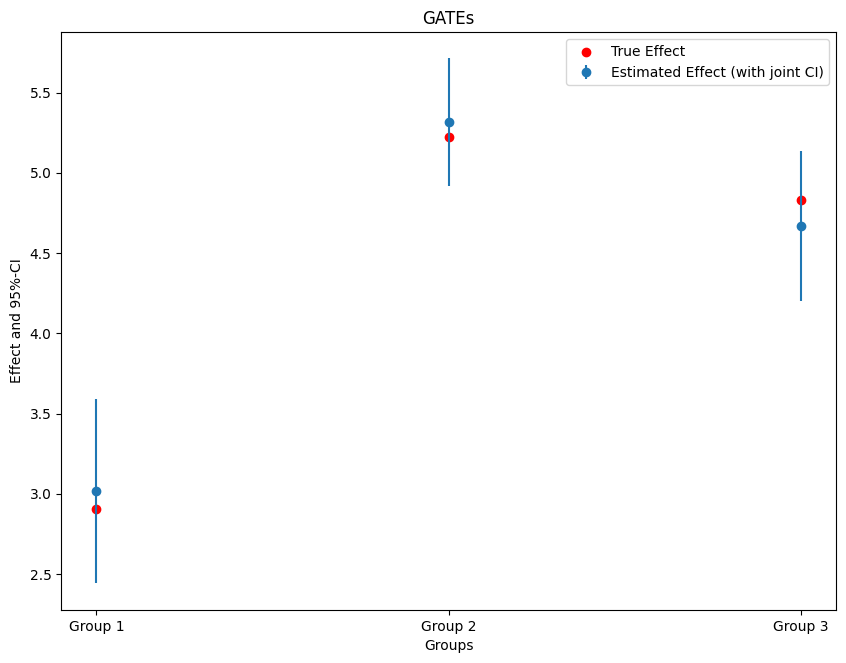
ci = gate.confint(level=0.95, joint=True, n_rep_boot=1000)
print(ci)
2.5 % effect 97.5 %
Group 1 2.441311 3.016702 3.592093
Group 2 4.917497 5.314781 5.712065
Group 3 4.200982 4.669562 5.138142
Finally, let us plot the estimates together with the true effect within each group.
[10]:
import matplotlib.pyplot as plt
plt.rcParams['figure.figsize'] = 10., 7.5
errors = np.full((2, ci.shape[0]), np.nan)
errors[0, :] = ci['effect'] - ci['2.5 %']
errors[1, :] = ci['97.5 %'] - ci['effect']
plt.errorbar(ci.index, ci.effect, fmt='o', yerr=errors, label='Estimated Effect (with joint CI)')
#add true effect
ax = plt.subplot(1, 1, 1)
ax.scatter(x=['Group 1', 'Group 2', 'Group 3'], y=true_effects, c='red', label='True Effect')
plt.title('GATEs')
plt.xlabel('Groups')
plt.legend()
_ = plt.ylabel('Effect and 95%-CI')
It is also possible to supply disjoint groups as a single vector (still as a data frame). Remark the slightly different name.
[11]:
groups = pd.DataFrame(columns=['Group'], index=range(data['X_0'].shape[0]), dtype=str)
for i, x_i in enumerate(data['X_0']):
if x_i <= 0.3:
groups.loc[i, 'Group'] = '1'
elif (x_i > 0.3) & (x_i <= 0.7):
groups.loc[i, 'Group'] = '2'
else:
groups.loc[i, 'Group'] = '3'
print(groups.head())
Group
0 3
1 2
2 2
3 2
4 2
This time lets consider pointwise confidence intervals.
[12]:
gate = dml_irm.gate(groups=groups)
ci = gate.confint()
print(ci)
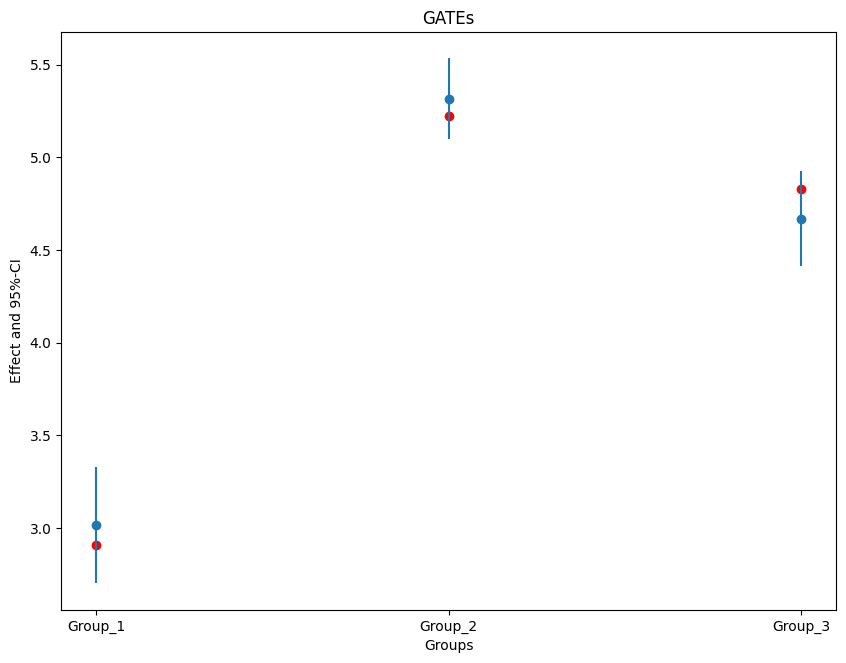
2.5 % effect 97.5 %
Group_1 2.701491 3.016702 3.331913
Group_2 5.097140 5.314781 5.532421
Group_3 4.412864 4.669562 4.926260
The coefficients of the best linear predictor can be seen via the summary (the values can be accessed through the underlying model .blp_model).
[13]:
print(gate.summary)
coef std err t P>|t| [0.025 0.975]
Group_1 3.016702 0.160825 18.757663 1.676093e-78 2.701491 3.331913
Group_2 5.314781 0.111043 47.862274 0.000000e+00 5.097140 5.532421
Group_3 4.669562 0.130971 35.653424 2.085279e-278 4.412864 4.926260
Remark that the confidence intervals in the summary are slightly smaller, since they are not based on the White’s heteroskedasticity robus standard errors.
[14]:
errors = np.full((2, ci.shape[0]), np.nan)
errors[0, :] = ci['effect'] - ci['2.5 %']
errors[1, :] = ci['97.5 %'] - ci['effect']
#add true effect
ax = plt.subplot(1, 1, 1)
ax.scatter(x=['Group_1', 'Group_2', 'Group_3'], y=true_effects, c='red', label='True Effect')
plt.errorbar(ci.index, ci.effect, fmt='o', yerr=errors)
plt.title('GATEs')
plt.xlabel('Groups')
_ = plt.ylabel('Effect and 95%-CI')