Note
-
Download Jupyter notebook:
https://docs.doubleml.org/stable/examples/R_double_ml_pipeline.ipynb.
R: Ensemble Learners and More with mlr3pipelines#
This notebook illustrates how to exploit the powerful tools provided by the mlr3pipelines package (Binder et al. 2021). For example, mlr3pipelines can be used in combination with DoubleML for feature engineering, combination of learners (ensemble learners, stacking), subsampling and hyperparameter tuning. The underlying idea of mlr3pipelines is to define a pipeline that incorporates a user’s desired operations. As a result, the pipeline returns an object of class Learner which can easily be passed to DoubleML. For an introduction to mlr3pipelines, we refer to the Pipelines Chapter in the mlr3book (Becker et al. 2020) and to the package website.
We intend to illustrate the major idea of how to use mlr3pipelines in combination with DoubleML in very simple examples. We use pipelines that are identical or very similar to the ones in the Pipelines Chapter in the mlr3book. Hence, we do not claim that the proposed learners are optimal in terms of their performance.
We start with the simple simulated data example and the Bonus data set from the Getting Started Section in the DoubleML user guide.
[1]:
library(DoubleML)
# Simulate data
set.seed(3141)
n_obs = 500
n_vars = 100
theta = 3
X = matrix(rnorm(n_obs*n_vars), nrow=n_obs, ncol=n_vars)
d = X[,1:3]%*%c(5,5,5) + rnorm(n_obs)
y = theta*d + X[, 1:3]%*%c(5,5,5) + rnorm(n_obs)
# Specify the data and variables for the causal model
# matrix interface to DoubleMLData
dml_data_sim = double_ml_data_from_matrix(X=X, y=y, d=d)
dml_data_sim
================= DoubleMLData Object ==================
------------------ Data summary ------------------
Outcome variable: y
Treatment variable(s): d
Covariates: X1, X2, X3, X4, X5, X6, X7, X8, X9, X10, X11, X12, X13, X14, X15, X16, X17, X18, X19, X20, X21, X22, X23, X24, X25, X26, X27, X28, X29, X30, X31, X32, X33, X34, X35, X36, X37, X38, X39, X40, X41, X42, X43, X44, X45, X46, X47, X48, X49, X50, X51, X52, X53, X54, X55, X56, X57, X58, X59, X60, X61, X62, X63, X64, X65, X66, X67, X68, X69, X70, X71, X72, X73, X74, X75, X76, X77, X78, X79, X80, X81, X82, X83, X84, X85, X86, X87, X88, X89, X90, X91, X92, X93, X94, X95, X96, X97, X98, X99, X100
Instrument(s):
No. Observations: 500
To have an example with a classification learner, we load the Bonus data set.
[2]:
# Load bonus data
df_bonus = fetch_bonus(return_type="data.table")
head(df_bonus)
# Specify the data and variables for the causal model
x_vars = c("female", "black", "othrace", "dep1", "dep2",
"q2", "q3", "q4", "q5", "q6", "agelt35", "agegt54",
"durable", "lusd", "husd")
dim_x = length(x_vars)
dml_data_bonus = DoubleMLData$new(df_bonus,
y_col = "inuidur1",
d_cols = "tg",
x_cols = x_vars)
print(dml_data_bonus)
| inuidur1 | female | black | othrace | dep1 | dep2 | q2 | q3 | q4 | q5 | q6 | agelt35 | agegt54 | durable | lusd | husd | tg |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> |
| 2.890372 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| 0.000000 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 3.295837 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 2.197225 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| 3.295837 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 |
| 3.295837 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 |
================= DoubleMLData Object ==================
------------------ Data summary ------------------
Outcome variable: inuidur1
Treatment variable(s): tg
Covariates: female, black, othrace, dep1, dep2, q2, q3, q4, q5, q6, agelt35, agegt54, durable, lusd, husd
Instrument(s):
No. Observations: 5099
To specify a learner for the nuisance part in a causal model, we can either use mlr3’s LearnerRegr for a model’s nuisance part with a continuous dependent variable or a LearnerClassif if the corresponding outcome variable is binary.
Moreover, it’s possible to create a learner based on a pipeline. For example, we could think of ensemble learners which combine several estimators.
Using learners from mlr3, mlr3learners and mlr3extralearners#
Let’s begin with a “standard” example on how to use any of the learners provided by mlr3 (Lang et al. 2020), mlr3learners (Lang et al. 2021) and mlr3extralearners (Sonabend and Schratz 2021) in DoubleML: We create an object of the class Learner which DoubleML internally uses for model training and generation of predictions.
In the simulated example, we will use a lasso estimator for the continuous treatment variable, which is based on the glmnet package. For the binary treatment variable in the Bonus data example, we use a random forest classifier as provided by ranger.
[3]:
library(mlr3)
library(mlr3learners)
# suppress messages during fitting
lgr::get_logger("mlr3")$set_threshold("warn")
learner_lasso = lrn("regr.cv_glmnet", s="lambda.min")
ml_l_lasso = learner_lasso$clone()
ml_m_lasso = learner_lasso$clone()
class(ml_l_lasso)
Warning message:
"Paket 'mlr3' wurde unter R Version 4.2.3 erstellt"
- 'LearnerRegrCVGlmnet'
- 'LearnerRegr'
- 'Learner'
- 'R6'
[4]:
# Random forest learner for nuisance part ml_l
learner_forest_regr = lrn("regr.ranger",
num.trees=500, mtry=floor(sqrt(dim_x)),
max.depth=5, min.node.size=2)
# Random forest learner for nuisance part ml_m (binary outcome)
learner_forest_classif = lrn("classif.ranger",
num.trees=500,
mtry=floor(sqrt(dim_x)),
max.depth=5, min.node.size=2)
ml_l_forest = learner_forest_regr$clone()
ml_m_forest = learner_forest_classif$clone()
class(ml_l_forest)
- 'LearnerRegrRanger'
- 'LearnerRegr'
- 'Learner'
- 'R6'
We set up a causal model, here we specify a partially linear model and thereby pass the learner as an input. Let’s fit the models.
[5]:
set.seed(123)
obj_dml_plr_sim = DoubleMLPLR$new(dml_data_sim,
ml_l=ml_l_lasso,
ml_m=ml_m_lasso)
obj_dml_plr_sim$fit()
print(obj_dml_plr_sim)
================= DoubleMLPLR Object ==================
------------------ Data summary ------------------
Outcome variable: y
Treatment variable(s): d
Covariates: X1, X2, X3, X4, X5, X6, X7, X8, X9, X10, X11, X12, X13, X14, X15, X16, X17, X18, X19, X20, X21, X22, X23, X24, X25, X26, X27, X28, X29, X30, X31, X32, X33, X34, X35, X36, X37, X38, X39, X40, X41, X42, X43, X44, X45, X46, X47, X48, X49, X50, X51, X52, X53, X54, X55, X56, X57, X58, X59, X60, X61, X62, X63, X64, X65, X66, X67, X68, X69, X70, X71, X72, X73, X74, X75, X76, X77, X78, X79, X80, X81, X82, X83, X84, X85, X86, X87, X88, X89, X90, X91, X92, X93, X94, X95, X96, X97, X98, X99, X100
Instrument(s):
No. Observations: 500
------------------ Score & algorithm ------------------
Score function: partialling out
DML algorithm: dml2
------------------ Machine learner ------------------
ml_l: regr.cv_glmnet
ml_m: regr.cv_glmnet
------------------ Resampling ------------------
No. folds: 5
No. repeated sample splits: 1
Apply cross-fitting: TRUE
------------------ Fit summary ------------------
Estimates and significance testing of the effect of target variables
Estimate. Std. Error t value Pr(>|t|)
d 3.01219 0.04415 68.22 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
[6]:
set.seed(123)
obj_dml_plr_bonus = DoubleMLPLR$new(dml_data_bonus,
ml_l=ml_l_forest,
ml_m=ml_m_forest)
obj_dml_plr_bonus$fit()
print(obj_dml_plr_bonus)
================= DoubleMLPLR Object ==================
------------------ Data summary ------------------
Outcome variable: inuidur1
Treatment variable(s): tg
Covariates: female, black, othrace, dep1, dep2, q2, q3, q4, q5, q6, agelt35, agegt54, durable, lusd, husd
Instrument(s):
No. Observations: 5099
------------------ Score & algorithm ------------------
Score function: partialling out
DML algorithm: dml2
------------------ Machine learner ------------------
ml_l: regr.ranger
ml_m: classif.ranger
------------------ Resampling ------------------
No. folds: 5
No. repeated sample splits: 1
Apply cross-fitting: TRUE
------------------ Fit summary ------------------
Estimates and significance testing of the effect of target variables
Estimate. Std. Error t value Pr(>|t|)
tg -0.0765 0.0354 -2.161 0.0307 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Set up learners based on mlr3pipelines#
These learners can also be constructed using mlr3pipelines. We’ll first use the PipeOp constructor po() to define the learner construction and then initiate a new instance of the Learner class. po() implements a computational step in a pipeline. For more information, we refer to the Pipelines Chapter in the mlr3book.
[7]:
# Lasso learner
library(mlr3pipelines)
pipe_lasso = po(lrn("regr.cv_glmnet"), s = "lambda.min")
ml_l_lasso_pipe = as_learner(pipe_lasso)
ml_m_lasso_pipe = as_learner(pipe_lasso)
# Class of the lasso learner
class(ml_l_lasso_pipe)
- 'GraphLearner'
- 'Learner'
- 'R6'
[8]:
# Random forest learner for nuisance part ml_l
pipe_forest_regr = po(lrn("regr.ranger"),
num.trees=500, mtry=floor(sqrt(dim_x)),
max.depth=5, min.node.size=2)
# Random forest learner for nuisance part ml_m (binary outcome)
pipe_forest_classif = po(lrn("classif.ranger"),
num.trees=500,
mtry=floor(sqrt(dim_x)),
max.depth=5, min.node.size=2)
ml_l_forest_pipe = as_learner(pipe_forest_regr)
ml_m_forest_pipe = as_learner(pipe_forest_classif)
# Class of the random forest learners
class(ml_l_forest_pipe)
class(ml_m_forest_pipe)
- 'GraphLearner'
- 'Learner'
- 'R6'
- 'GraphLearner'
- 'Learner'
- 'R6'
Let’s use these learners to fit the PLR in both examples.
[9]:
set.seed(123)
obj_dml_plr_sim_pipe = DoubleMLPLR$new(dml_data_sim,
ml_l=ml_l_lasso_pipe,
ml_m=ml_m_lasso_pipe)
obj_dml_plr_sim_pipe$fit()
print(obj_dml_plr_sim_pipe)
================= DoubleMLPLR Object ==================
------------------ Data summary ------------------
Outcome variable: y
Treatment variable(s): d
Covariates: X1, X2, X3, X4, X5, X6, X7, X8, X9, X10, X11, X12, X13, X14, X15, X16, X17, X18, X19, X20, X21, X22, X23, X24, X25, X26, X27, X28, X29, X30, X31, X32, X33, X34, X35, X36, X37, X38, X39, X40, X41, X42, X43, X44, X45, X46, X47, X48, X49, X50, X51, X52, X53, X54, X55, X56, X57, X58, X59, X60, X61, X62, X63, X64, X65, X66, X67, X68, X69, X70, X71, X72, X73, X74, X75, X76, X77, X78, X79, X80, X81, X82, X83, X84, X85, X86, X87, X88, X89, X90, X91, X92, X93, X94, X95, X96, X97, X98, X99, X100
Instrument(s):
No. Observations: 500
------------------ Score & algorithm ------------------
Score function: partialling out
DML algorithm: dml2
------------------ Machine learner ------------------
ml_l: regr.cv_glmnet
ml_m: regr.cv_glmnet
------------------ Resampling ------------------
No. folds: 5
No. repeated sample splits: 1
Apply cross-fitting: TRUE
------------------ Fit summary ------------------
Estimates and significance testing of the effect of target variables
Estimate. Std. Error t value Pr(>|t|)
d 3.01219 0.04415 68.22 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
[10]:
set.seed(123)
obj_dml_plr_bonus_pipe = DoubleMLPLR$new(dml_data_bonus,
ml_l=ml_l_forest_pipe,
ml_m=ml_m_forest_pipe)
obj_dml_plr_bonus_pipe$fit()
print(obj_dml_plr_bonus_pipe)
================= DoubleMLPLR Object ==================
------------------ Data summary ------------------
Outcome variable: inuidur1
Treatment variable(s): tg
Covariates: female, black, othrace, dep1, dep2, q2, q3, q4, q5, q6, agelt35, agegt54, durable, lusd, husd
Instrument(s):
No. Observations: 5099
------------------ Score & algorithm ------------------
Score function: partialling out
DML algorithm: dml2
------------------ Machine learner ------------------
ml_l: regr.ranger
ml_m: classif.ranger
------------------ Resampling ------------------
No. folds: 5
No. repeated sample splits: 1
Apply cross-fitting: TRUE
------------------ Fit summary ------------------
Estimates and significance testing of the effect of target variables
Estimate. Std. Error t value Pr(>|t|)
tg -0.0765 0.0354 -2.161 0.0307 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Use ensemble learners based on mlr3pipelines#
First, let’s see how we can use more complicated GraphLearners like ensemble learners in DoubleML. For example, we want to create a learner with predictions that are generated as an average from three different learners. In the first step, we split up the pipeline into three branches. In our example, the learners estimate the nuisance parts independently of each other. In the last step,
we average the predictions. Thereby we will use the pipe operator %>>%. For more details, we refer to the Pipelines Chapter in the mlr3book.
[11]:
# For regression (nuisance parts with continuous outcome)
graph_ensemble_regr = gunion(list(
po("learner", lrn("regr.cv_glmnet", s = "lambda.min")),
po("learner", lrn("regr.ranger")),
po("learner", lrn("regr.rpart", cp = 0.01))
)) %>>%
po("regravg", 3)
# Class of ' graph_ensemble_regr'
class(graph_ensemble_regr)
- 'Graph'
- 'R6'
[12]:
# Plot the graph
graph_ensemble_regr$plot()
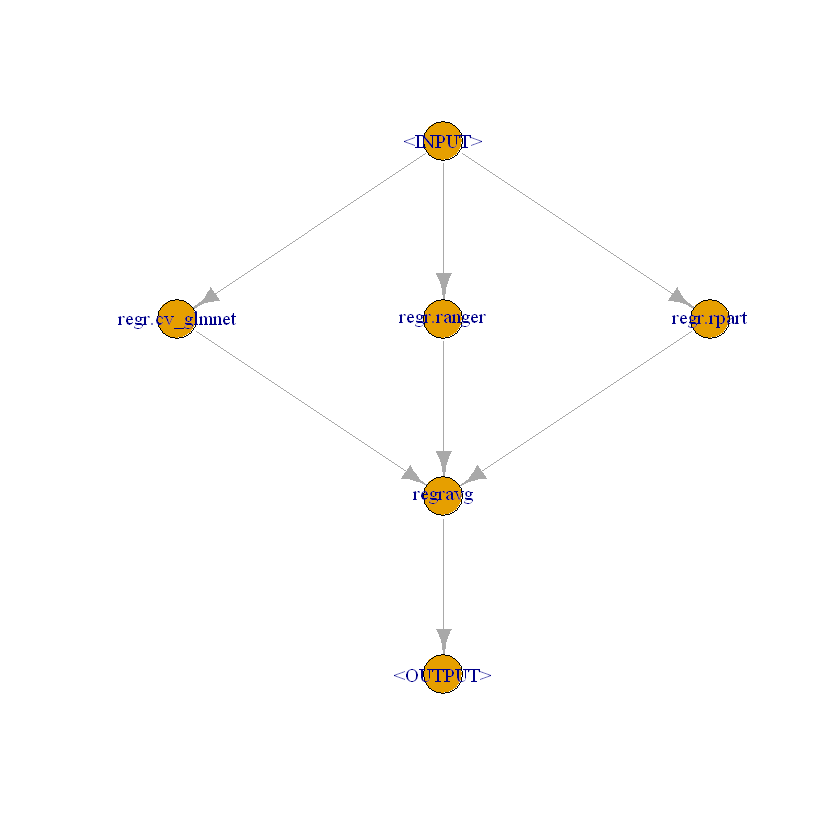
[13]:
# For classification (nuisance part ml_m in the Bonus example)
graph_ensemble_classif = gunion(list(
po("learner", lrn("classif.cv_glmnet", s = "lambda.min")),
po("learner", lrn("classif.ranger")),
po("learner", lrn("classif.rpart", cp = 0.01))
)) %>>%
po("classifavg", 3)
# Class of 'graph_ensemble_classif'
class(graph_ensemble_classif)
- 'Graph'
- 'R6'
[14]:
# Plot the graph
graph_ensemble_classif$plot()
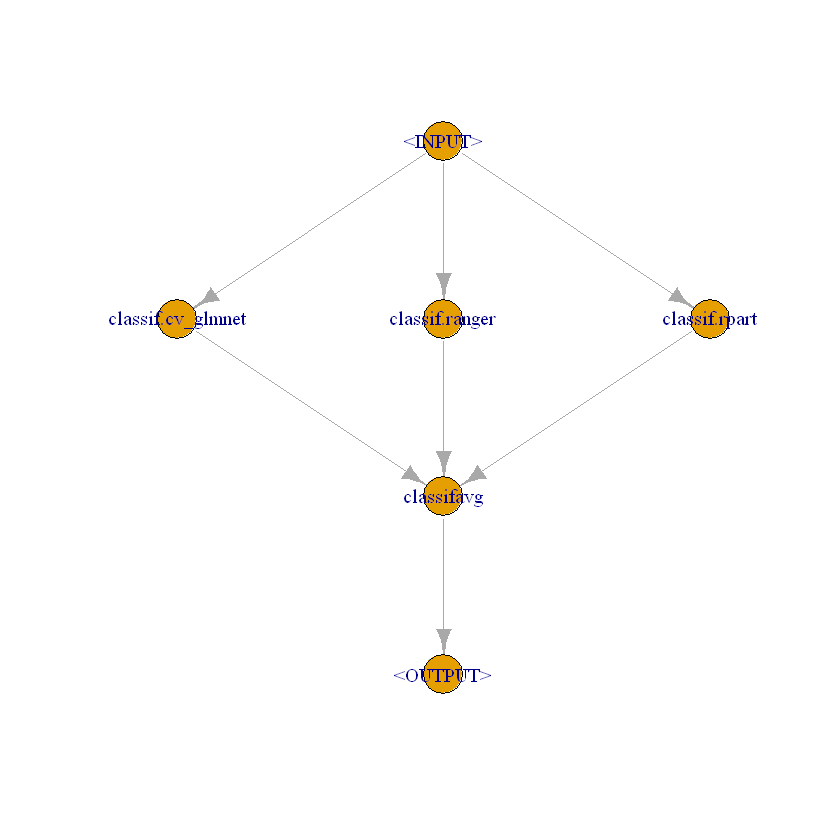
We create a new instance of a GraphLearner which is later used in DoubleML.
[15]:
ensemble_pipe_regr = as_learner(graph_ensemble_regr)
ensemble_pipe_classif = as_learner(graph_ensemble_classif)
Let’s estimate the two PLR examples with the ensemble learner.
[16]:
# Initiate new DoubleML object and estimate with graph learner
set.seed(123)
obj_dml_plr_sim_pipe_ensemble = DoubleMLPLR$new(dml_data_sim,
ml_l = ensemble_pipe_regr,
ml_m = ensemble_pipe_regr)
obj_dml_plr_sim_pipe_ensemble$fit()
print(obj_dml_plr_sim_pipe_ensemble)
================= DoubleMLPLR Object ==================
------------------ Data summary ------------------
Outcome variable: y
Treatment variable(s): d
Covariates: X1, X2, X3, X4, X5, X6, X7, X8, X9, X10, X11, X12, X13, X14, X15, X16, X17, X18, X19, X20, X21, X22, X23, X24, X25, X26, X27, X28, X29, X30, X31, X32, X33, X34, X35, X36, X37, X38, X39, X40, X41, X42, X43, X44, X45, X46, X47, X48, X49, X50, X51, X52, X53, X54, X55, X56, X57, X58, X59, X60, X61, X62, X63, X64, X65, X66, X67, X68, X69, X70, X71, X72, X73, X74, X75, X76, X77, X78, X79, X80, X81, X82, X83, X84, X85, X86, X87, X88, X89, X90, X91, X92, X93, X94, X95, X96, X97, X98, X99, X100
Instrument(s):
No. Observations: 500
------------------ Score & algorithm ------------------
Score function: partialling out
DML algorithm: dml2
------------------ Machine learner ------------------
ml_l: regr.cv_glmnet.regr.ranger.regr.rpart.regravg
ml_m: regr.cv_glmnet.regr.ranger.regr.rpart.regravg
------------------ Resampling ------------------
No. folds: 5
No. repeated sample splits: 1
Apply cross-fitting: TRUE
------------------ Fit summary ------------------
Estimates and significance testing of the effect of target variables
Estimate. Std. Error t value Pr(>|t|)
d 3.88664 0.02584 150.4 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
[17]:
set.seed(123)
obj_dml_plr_bonus_pipe_ensemble = DoubleMLPLR$new(dml_data_bonus,
ml_l = ensemble_pipe_regr,
ml_m = ensemble_pipe_classif)
obj_dml_plr_bonus_pipe_ensemble$fit()
print(obj_dml_plr_bonus_pipe_ensemble)
================= DoubleMLPLR Object ==================
------------------ Data summary ------------------
Outcome variable: inuidur1
Treatment variable(s): tg
Covariates: female, black, othrace, dep1, dep2, q2, q3, q4, q5, q6, agelt35, agegt54, durable, lusd, husd
Instrument(s):
No. Observations: 5099
------------------ Score & algorithm ------------------
Score function: partialling out
DML algorithm: dml2
------------------ Machine learner ------------------
ml_l: regr.cv_glmnet.regr.ranger.regr.rpart.regravg
ml_m: classif.cv_glmnet.classif.ranger.classif.rpart.classifavg
------------------ Resampling ------------------
No. folds: 5
No. repeated sample splits: 1
Apply cross-fitting: TRUE
------------------ Fit summary ------------------
Estimates and significance testing of the effect of target variables
Estimate. Std. Error t value Pr(>|t|)
tg -0.07689 0.03545 -2.169 0.0301 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Alternatively, different learners could also be stacked. Here we simply repeat the example from the Pipelines Chapter in the mlr3book in our Bonus data example.
[18]:
lrn = lrn("classif.rpart")
lrn_0 = po("learner_cv", lrn$clone())
lrn_0$id = "rpart_cv"
[19]:
# Pass original features to final estimation step
level_0 = gunion(list(lrn_0, po("nop")))
[20]:
combined = level_0 %>>% po("featureunion", 2)
[21]:
stack = combined %>>% po("learner", lrn$clone())
stack$plot(html = FALSE)
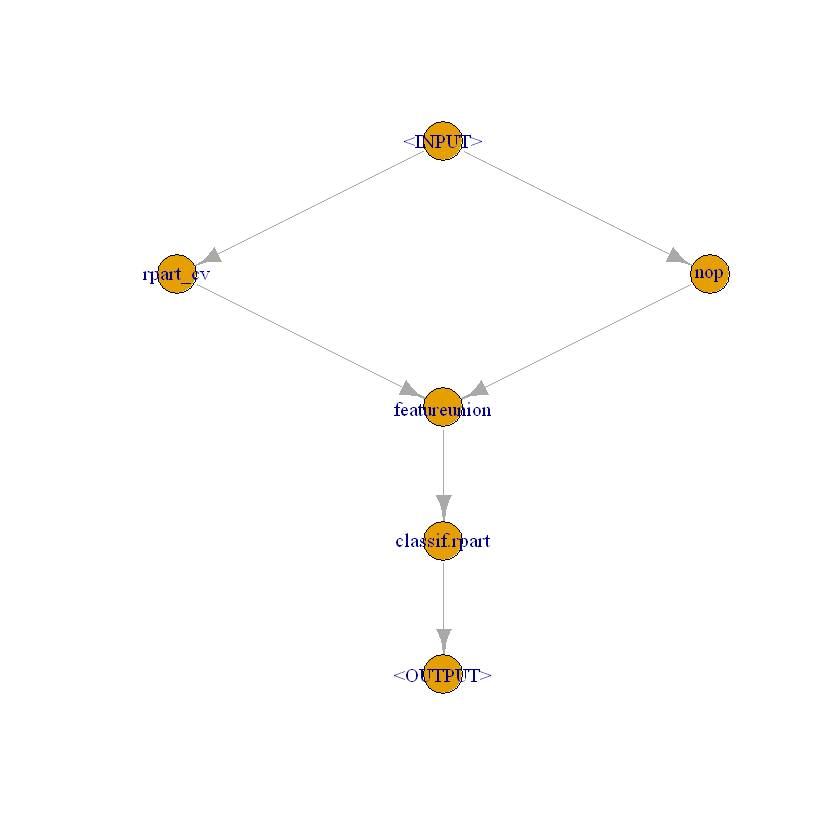
[22]:
# Create a stacked learner and pass it to a DoubleML object
stacklrn = as_learner(stack)
set.seed(123)
obj_dml_plr_bonus_pipe = DoubleMLPLR$new(dml_data_bonus,
ml_l=ml_l_forest,
ml_m=stacklrn)
obj_dml_plr_bonus_pipe$fit()
print(obj_dml_plr_bonus_pipe)
================= DoubleMLPLR Object ==================
------------------ Data summary ------------------
Outcome variable: inuidur1
Treatment variable(s): tg
Covariates: female, black, othrace, dep1, dep2, q2, q3, q4, q5, q6, agelt35, agegt54, durable, lusd, husd
Instrument(s):
No. Observations: 5099
------------------ Score & algorithm ------------------
Score function: partialling out
DML algorithm: dml2
------------------ Machine learner ------------------
ml_l: regr.ranger
ml_m: rpart_cv.nop.featureunion.classif.rpart
------------------ Resampling ------------------
No. folds: 5
No. repeated sample splits: 1
Apply cross-fitting: TRUE
------------------ Fit summary ------------------
Estimates and significance testing of the effect of target variables
Estimate. Std. Error t value Pr(>|t|)
tg -0.07915 0.03538 -2.237 0.0253 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
How to exploit more features of mlr3pipelines in DoubleML#
mlr3pipelines can do much more. For example, we could use it to perform some feature engineering and even perform pipeline-based parameter tuning. We just have to define the steps we want to have in our pipeline by using the PipeOps.
Let’s have a look at two more examples from the Pipelines Chapter in the mlr3book. In the first one, we will do some data manipulation. The second example illustrate how we could use mlr3pipelines for parameter tuning.
Data preprocessing#
Let’s perform some data preprocessing and then use a regression tree for prediction.
[23]:
mutate = po("mutate")
filter = po("filter",
filter = mlr3filters::flt("variance"),
param_vals = list(filter.frac = 0.5))
Collect them in a graph and plot it.
[24]:
graph = mutate %>>%
filter %>>%
po("learner",
learner = lrn("classif.rpart"))
class(graph)
- 'Graph'
- 'R6'
[25]:
graph$plot()
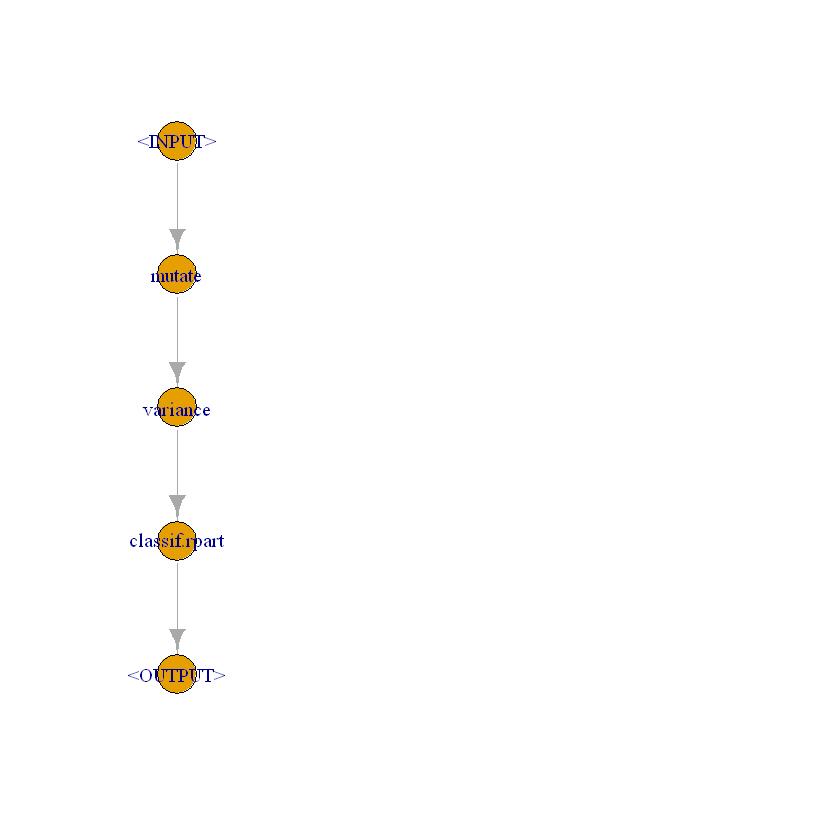
Create a new learner.
[26]:
glrn = as_learner(graph)
[27]:
glrn
class(glrn)
<GraphLearner:mutate.variance.classif.rpart>
* Model: -
* Parameters: mutate.mutation=<list>, mutate.delete_originals=FALSE,
variance.filter.frac=0.5, classif.rpart.xval=0
* Packages: mlr3, mlr3pipelines, rpart
* Predict Types: [response], prob
* Feature Types: logical, integer, numeric, character, factor, ordered,
POSIXct
* Properties: featureless, hotstart_backward, hotstart_forward,
importance, loglik, missings, multiclass, oob_error,
selected_features, twoclass, weights
- 'GraphLearner'
- 'Learner'
- 'R6'
[28]:
set.seed(123)
obj_dml_plr_bonus_pipe2 = DoubleMLPLR$new(dml_data_bonus,
ml_l=ml_l_lasso,
ml_m=glrn)
obj_dml_plr_bonus_pipe2$fit()
print(obj_dml_plr_bonus_pipe2)
================= DoubleMLPLR Object ==================
------------------ Data summary ------------------
Outcome variable: inuidur1
Treatment variable(s): tg
Covariates: female, black, othrace, dep1, dep2, q2, q3, q4, q5, q6, agelt35, agegt54, durable, lusd, husd
Instrument(s):
No. Observations: 5099
------------------ Score & algorithm ------------------
Score function: partialling out
DML algorithm: dml2
------------------ Machine learner ------------------
ml_l: regr.cv_glmnet
ml_m: mutate.variance.classif.rpart
------------------ Resampling ------------------
No. folds: 5
No. repeated sample splits: 1
Apply cross-fitting: TRUE
------------------ Fit summary ------------------
Estimates and significance testing of the effect of target variables
Estimate. Std. Error t value Pr(>|t|)
tg -0.07366 0.03539 -2.081 0.0374 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Let’s see how to set hyperparameters with a pipeline.
[29]:
glrn$param_set$values$variance.filter.frac = 0.25
[30]:
set.seed(123)
obj_dml_plr_bonus_pipe3 = DoubleMLPLR$new(dml_data_bonus,
ml_l=ml_l_lasso,
ml_m=glrn)
obj_dml_plr_bonus_pipe3$fit()
print(obj_dml_plr_bonus_pipe3)
================= DoubleMLPLR Object ==================
------------------ Data summary ------------------
Outcome variable: inuidur1
Treatment variable(s): tg
Covariates: female, black, othrace, dep1, dep2, q2, q3, q4, q5, q6, agelt35, agegt54, durable, lusd, husd
Instrument(s):
No. Observations: 5099
------------------ Score & algorithm ------------------
Score function: partialling out
DML algorithm: dml2
------------------ Machine learner ------------------
ml_l: regr.cv_glmnet
ml_m: mutate.variance.classif.rpart
------------------ Resampling ------------------
No. folds: 5
No. repeated sample splits: 1
Apply cross-fitting: TRUE
------------------ Fit summary ------------------
Estimates and significance testing of the effect of target variables
Estimate. Std. Error t value Pr(>|t|)
tg -0.07366 0.03539 -2.081 0.0374 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Parameter tuning#
Next, we will shortly illustrate how to perform parameter tuning in the simulated data example. Here, we use a pipeline to tune the penalty of the lasso. To do so, we generate a GraphLearner and then call DoubleML’s tune() method.
Let’s define a GraphLearner based on the lasso.
[31]:
lasso_pipe = mutate %>>%
filter %>>%
po("learner",
learner = lrn("regr.glmnet"))
glrn_lasso = as_learner(lasso_pipe)
Let’s specify the parameter grid and more settings that are required for the parameter tuning. For more details, we refer to the DoubleML user guide.
[32]:
# Parameter grid for lambda and for optimal variance filter fraction
library(paradox)
par_grids = ps(regr.glmnet.lambda = p_dbl(lower = 0.05, upper = 0.1),
variance.filter.frac = p_dbl(lower = 0.25, upper = 1))
Warning message:
"Paket 'paradox' wurde unter R Version 4.2.3 erstellt"
[33]:
# Specify further tune settings
library(mlr3tuning)
tune_settings = list(terminator = trm("evals", n_evals = 10),
algorithm = tnr("grid_search", resolution = 10),
rsmp_tune = rsmp("cv", folds = 5),
measure = list("ml_l" = msr("regr.mse"),
"ml_m" = msr("regr.mse")))
[34]:
# Initiate new DoubleML object and execute tuning with graph learner
set.seed(123)
obj_dml_plr_sim_pipe_tune = DoubleMLPLR$new(dml_data_sim,
ml_l=glrn_lasso,
ml_m=glrn_lasso)
obj_dml_plr_sim_pipe_tune$tune(param_set = list("ml_l" = par_grids,
"ml_m" = par_grids),
tune_settings=tune_settings)
INFO [12:57:48.469] [bbotk] Starting to optimize 2 parameter(s) with '<TunerGridSearch>' and '<TerminatorEvals> [n_evals=10, k=0]'
INFO [12:57:48.485] [bbotk] Evaluating 1 configuration(s)
INFO [12:57:49.133] [bbotk] Result of batch 1:
INFO [12:57:49.135] [bbotk] regr.glmnet.lambda variance.filter.frac regr.mse warnings errors
INFO [12:57:49.135] [bbotk] 0.07222222 0.6666667 107.2925 0 0
INFO [12:57:49.135] [bbotk] runtime_learners uhash
INFO [12:57:49.135] [bbotk] 0.38 5574dcd4-fc9e-463b-9345-d21ee5775b5f
INFO [12:57:49.137] [bbotk] Evaluating 1 configuration(s)
INFO [12:57:49.514] [bbotk] Result of batch 2:
INFO [12:57:49.516] [bbotk] regr.glmnet.lambda variance.filter.frac regr.mse warnings errors
INFO [12:57:49.516] [bbotk] 0.1 0.6666667 106.2652 0 0
INFO [12:57:49.516] [bbotk] runtime_learners uhash
INFO [12:57:49.516] [bbotk] 0.32 8497f641-2700-4a53-ae89-5cb31a99b9cc
INFO [12:57:49.517] [bbotk] Evaluating 1 configuration(s)
INFO [12:57:49.892] [bbotk] Result of batch 3:
INFO [12:57:49.893] [bbotk] regr.glmnet.lambda variance.filter.frac regr.mse warnings errors
INFO [12:57:49.893] [bbotk] 0.1 0.4166667 482.9753 0 0
INFO [12:57:49.893] [bbotk] runtime_learners uhash
INFO [12:57:49.893] [bbotk] 0.33 f3d24993-a09b-432f-ab71-44fa97767be8
INFO [12:57:49.894] [bbotk] Evaluating 1 configuration(s)
INFO [12:57:50.273] [bbotk] Result of batch 4:
INFO [12:57:50.274] [bbotk] regr.glmnet.lambda variance.filter.frac regr.mse warnings errors
INFO [12:57:50.274] [bbotk] 0.09444444 0.75 112.1054 0 0
INFO [12:57:50.274] [bbotk] runtime_learners uhash
INFO [12:57:50.274] [bbotk] 0.3 bb2913dc-3cd0-4b8f-a09a-303f00f0bd62
INFO [12:57:50.276] [bbotk] Evaluating 1 configuration(s)
INFO [12:57:50.657] [bbotk] Result of batch 5:
INFO [12:57:50.659] [bbotk] regr.glmnet.lambda variance.filter.frac regr.mse warnings errors
INFO [12:57:50.659] [bbotk] 0.08333333 0.9166667 10.81856 0 0
INFO [12:57:50.659] [bbotk] runtime_learners uhash
INFO [12:57:50.659] [bbotk] 0.28 8da924ce-f2e7-4dd2-a840-b5d34a6f42be
INFO [12:57:50.660] [bbotk] Evaluating 1 configuration(s)
INFO [12:57:51.051] [bbotk] Result of batch 6:
INFO [12:57:51.053] [bbotk] regr.glmnet.lambda variance.filter.frac regr.mse warnings errors
INFO [12:57:51.053] [bbotk] 0.08888889 0.9166667 10.74189 0 0
INFO [12:57:51.053] [bbotk] runtime_learners uhash
INFO [12:57:51.053] [bbotk] 0.29 0434e374-ddc9-468d-b208-dc13a11076b3
INFO [12:57:51.054] [bbotk] Evaluating 1 configuration(s)
INFO [12:57:51.433] [bbotk] Result of batch 7:
INFO [12:57:51.435] [bbotk] regr.glmnet.lambda variance.filter.frac regr.mse warnings errors
INFO [12:57:51.435] [bbotk] 0.07777778 0.5833333 201.8513 0 0
INFO [12:57:51.435] [bbotk] runtime_learners uhash
INFO [12:57:51.435] [bbotk] 0.26 280454dd-5804-498f-80a8-24080030a4de
INFO [12:57:51.436] [bbotk] Evaluating 1 configuration(s)
INFO [12:57:51.868] [bbotk] Result of batch 8:
INFO [12:57:51.869] [bbotk] regr.glmnet.lambda variance.filter.frac regr.mse warnings errors
INFO [12:57:51.869] [bbotk] 0.07222222 0.3333333 531.5255 0 0
INFO [12:57:51.869] [bbotk] runtime_learners uhash
INFO [12:57:51.869] [bbotk] 0.32 7b428990-305b-47be-bde4-7215093d9089
INFO [12:57:51.871] [bbotk] Evaluating 1 configuration(s)
INFO [12:57:52.316] [bbotk] Result of batch 9:
INFO [12:57:52.318] [bbotk] regr.glmnet.lambda variance.filter.frac regr.mse warnings errors
INFO [12:57:52.318] [bbotk] 0.06111111 0.9166667 11.17823 0 0
INFO [12:57:52.318] [bbotk] runtime_learners uhash
INFO [12:57:52.318] [bbotk] 0.34 bd929a9e-3e1c-4fee-ae56-f00584a57972
INFO [12:57:52.319] [bbotk] Evaluating 1 configuration(s)
INFO [12:57:52.773] [bbotk] Result of batch 10:
INFO [12:57:52.775] [bbotk] regr.glmnet.lambda variance.filter.frac regr.mse warnings errors
INFO [12:57:52.775] [bbotk] 0.08888889 0.25 538.991 0 0
INFO [12:57:52.775] [bbotk] runtime_learners uhash
INFO [12:57:52.775] [bbotk] 0.38 e20ea26e-a6ba-4539-a3d9-0005a80b528f
INFO [12:57:52.780] [bbotk] Finished optimizing after 10 evaluation(s)
INFO [12:57:52.780] [bbotk] Result:
INFO [12:57:52.782] [bbotk] regr.glmnet.lambda variance.filter.frac learner_param_vals x_domain regr.mse
INFO [12:57:52.782] [bbotk] 0.08888889 0.9166667 <list[5]> <list[2]> 10.74189
INFO [12:57:52.869] [bbotk] Starting to optimize 2 parameter(s) with '<TunerGridSearch>' and '<TerminatorEvals> [n_evals=10, k=0]'
INFO [12:57:52.871] [bbotk] Evaluating 1 configuration(s)
INFO [12:57:53.298] [bbotk] Result of batch 1:
INFO [12:57:53.299] [bbotk] regr.glmnet.lambda variance.filter.frac regr.mse warnings errors
INFO [12:57:53.299] [bbotk] 0.08888889 0.5833333 12.6173 0 0
INFO [12:57:53.299] [bbotk] runtime_learners uhash
INFO [12:57:53.299] [bbotk] 0.35 67ad635a-5346-41e5-b5d7-a79359d2da46
INFO [12:57:53.301] [bbotk] Evaluating 1 configuration(s)
INFO [12:57:53.723] [bbotk] Result of batch 2:
INFO [12:57:53.725] [bbotk] regr.glmnet.lambda variance.filter.frac regr.mse warnings errors
INFO [12:57:53.725] [bbotk] 0.1 0.5833333 12.5155 0 0
INFO [12:57:53.725] [bbotk] runtime_learners uhash
INFO [12:57:53.725] [bbotk] 0.34 921e4f0d-e57c-4552-a5e6-8bdee1a1d83d
INFO [12:57:53.726] [bbotk] Evaluating 1 configuration(s)
INFO [12:57:54.130] [bbotk] Result of batch 3:
INFO [12:57:54.132] [bbotk] regr.glmnet.lambda variance.filter.frac regr.mse warnings errors
INFO [12:57:54.132] [bbotk] 0.08333333 0.75 0.9870004 0 0
INFO [12:57:54.132] [bbotk] runtime_learners uhash
INFO [12:57:54.132] [bbotk] 0.3 26bd56a6-fd8a-4dba-9109-0ff823b17d45
INFO [12:57:54.133] [bbotk] Evaluating 1 configuration(s)
INFO [12:57:54.574] [bbotk] Result of batch 4:
INFO [12:57:54.576] [bbotk] regr.glmnet.lambda variance.filter.frac regr.mse warnings errors
INFO [12:57:54.576] [bbotk] 0.07222222 0.5 17.75887 0 0
INFO [12:57:54.576] [bbotk] runtime_learners uhash
INFO [12:57:54.576] [bbotk] 0.34 fb5c25fa-1596-49d4-b371-d0cdb0ea4795
INFO [12:57:54.577] [bbotk] Evaluating 1 configuration(s)
INFO [12:57:55.015] [bbotk] Result of batch 5:
INFO [12:57:55.017] [bbotk] regr.glmnet.lambda variance.filter.frac regr.mse warnings errors
INFO [12:57:55.017] [bbotk] 0.08333333 0.5833333 12.6722 0 0
INFO [12:57:55.017] [bbotk] runtime_learners uhash
INFO [12:57:55.017] [bbotk] 0.33 ee97bda7-6cea-440a-9248-d5a0c70f1d98
INFO [12:57:55.018] [bbotk] Evaluating 1 configuration(s)
INFO [12:57:55.452] [bbotk] Result of batch 6:
INFO [12:57:55.453] [bbotk] regr.glmnet.lambda variance.filter.frac regr.mse warnings errors
INFO [12:57:55.453] [bbotk] 0.07222222 0.5833333 12.78818 0 0
INFO [12:57:55.453] [bbotk] runtime_learners uhash
INFO [12:57:55.453] [bbotk] 0.29 8e3aa840-c895-472e-85c5-681817dcfcda
INFO [12:57:55.455] [bbotk] Evaluating 1 configuration(s)
INFO [12:57:55.913] [bbotk] Result of batch 7:
INFO [12:57:55.915] [bbotk] regr.glmnet.lambda variance.filter.frac regr.mse warnings errors
INFO [12:57:55.915] [bbotk] 0.07777778 0.5 17.69921 0 0
INFO [12:57:55.915] [bbotk] runtime_learners uhash
INFO [12:57:55.915] [bbotk] 0.36 4552b8af-3647-43f0-a5e7-55dc37e31fb1
INFO [12:57:55.917] [bbotk] Evaluating 1 configuration(s)
INFO [12:57:56.386] [bbotk] Result of batch 8:
INFO [12:57:56.387] [bbotk] regr.glmnet.lambda variance.filter.frac regr.mse warnings errors
INFO [12:57:56.387] [bbotk] 0.07777778 0.9166667 0.9880384 0 0
INFO [12:57:56.387] [bbotk] runtime_learners uhash
INFO [12:57:56.387] [bbotk] 0.31 abb0fd28-0359-4ecd-8644-f1718fdeb9b0
INFO [12:57:56.389] [bbotk] Evaluating 1 configuration(s)
INFO [12:57:56.783] [bbotk] Result of batch 9:
INFO [12:57:56.785] [bbotk] regr.glmnet.lambda variance.filter.frac regr.mse warnings errors
INFO [12:57:56.785] [bbotk] 0.07777778 0.4166667 22.43294 0 0
INFO [12:57:56.785] [bbotk] runtime_learners uhash
INFO [12:57:56.785] [bbotk] 0.26 cda85647-3ec2-4849-88ad-193f0d909729
INFO [12:57:56.786] [bbotk] Evaluating 1 configuration(s)
INFO [12:57:57.183] [bbotk] Result of batch 10:
INFO [12:57:57.185] [bbotk] regr.glmnet.lambda variance.filter.frac regr.mse warnings errors
INFO [12:57:57.185] [bbotk] 0.07222222 0.3333333 27.31378 0 0
INFO [12:57:57.185] [bbotk] runtime_learners uhash
INFO [12:57:57.185] [bbotk] 0.31 caac5a95-4462-42ba-99c8-ca1af7be64b2
INFO [12:57:57.189] [bbotk] Finished optimizing after 10 evaluation(s)
INFO [12:57:57.190] [bbotk] Result:
INFO [12:57:57.191] [bbotk] regr.glmnet.lambda variance.filter.frac learner_param_vals x_domain regr.mse
INFO [12:57:57.191] [bbotk] 0.08333333 0.75 <list[5]> <list[2]> 0.9870004
[35]:
obj_dml_plr_sim_pipe$fit()
print(obj_dml_plr_sim_pipe)
================= DoubleMLPLR Object ==================
------------------ Data summary ------------------
Outcome variable: y
Treatment variable(s): d
Covariates: X1, X2, X3, X4, X5, X6, X7, X8, X9, X10, X11, X12, X13, X14, X15, X16, X17, X18, X19, X20, X21, X22, X23, X24, X25, X26, X27, X28, X29, X30, X31, X32, X33, X34, X35, X36, X37, X38, X39, X40, X41, X42, X43, X44, X45, X46, X47, X48, X49, X50, X51, X52, X53, X54, X55, X56, X57, X58, X59, X60, X61, X62, X63, X64, X65, X66, X67, X68, X69, X70, X71, X72, X73, X74, X75, X76, X77, X78, X79, X80, X81, X82, X83, X84, X85, X86, X87, X88, X89, X90, X91, X92, X93, X94, X95, X96, X97, X98, X99, X100
Instrument(s):
No. Observations: 500
------------------ Score & algorithm ------------------
Score function: partialling out
DML algorithm: dml2
------------------ Machine learner ------------------
ml_l: regr.cv_glmnet
ml_m: regr.cv_glmnet
------------------ Resampling ------------------
No. folds: 5
No. repeated sample splits: 1
Apply cross-fitting: TRUE
------------------ Fit summary ------------------
Estimates and significance testing of the effect of target variables
Estimate. Std. Error t value Pr(>|t|)
d 3.00133 0.04424 67.84 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
References#
Becker, M., Binder, M., Bischl, B., Lang, M., Pfisterer, F., Reich, N.G., Richter, J., Schratz, P., Sonabend, R. (2020), mlr3 book, available at https://mlr3book.mlr-org.com.
Binder, M., Pfisterer, F., Lang, M., Schneider, L., Kotthof, L., and Bischl, B. (2021), mlr3pipelines - flexible machine learning pipelines in R, Journal of Machine Learning Research, 22(184): 1-7, https://jmlr.org/papers/v22/21-0281.html.
Lang, M., Binder, M., Richter, J., Schratz, P., Pfisterer, F., Coors, S., Au, Q., Casalicchio, G., Kotthoff, L., Bischl, B. (2019), mlr3: A modern object-oriented machine learing framework in R. Journal of Open Source Software, doi:10.21105/joss.01903.
Lang, M., Au, Q., Coors, S., and Schratz, P. (2021), mlr3learners: Recommended learners for mlr3, R package, https://CRAN.R-project.org/package=mlr3learners.
Sonabend, R., and Schratz, P. (2021), extralearners: Extra learners for mlr3. R package, https://mlr3extralearners.mlr-org.com/.
Acknowledgement
We would like to thank the developers of the mlr3pipelines package for providing such a powerful and easy-to-use implementation of many important ML tools.

