| coef | std err | t | P>|t| | 2.5 % | 97.5 % | |
|---|---|---|---|---|---|---|
| e401 | 8121.565 | 1106.553 | 7.34 | 0.0 | 5952.76 | 10290.369 |
DoubleML - Sensitivity Analysis
Tools for Causality
Grenoble, Sept 25 - 29, 2023
Philipp Bach, Sven Klaassen
Motivation: Sensitivity Analysis
Motivation: DoubleML Workflow
Problem Formulation
Data-Backend
Causal Model
ML Methods
DML Specification
Estimation
Inference
7. Sensitivity Analysis
Earlier we introduced 6 steps of the DoubleML workflow.
Actually, there is a 7th step that is often overlooked: Sensitivity Analysis
Motivation
- Whenever we have (properly collected) experimental data, we have good reason to believe that the independence assumption holds, i.e., the treatment assignment is independent of the potential outcomes
\[ Y(d) ⊥ D \]
- However, what about causal evidence from observational data? 😱
Motivation
Observational studies are often based on the assumption of conditional independence1
Conditional on pre-treatment confounders \(X\), the treatment is as good as randomly assigned
\[ Y(d) ⊥ D \mid X \]
- In general, the independence assumption is not testable!
Code
import networkx as nx
import matplotlib.pyplot as plt
G = nx.DiGraph()
# Add nodes
G.add_node("D")
G.add_node("Y")
G.add_node("X")
G.add_edge("D", "Y")
G.add_edge("X", "Y")
G.add_edge("X", "D")
# Draw the graph
plt.figure(figsize=(4, 3))
pos = {"D": (0, 0), "Y": (2, 0), "X": (1,1)}
edge_colors = ['black', 'black', 'black']
nx.draw(G, pos, with_labels=True, node_size=800, node_color='lightblue',
edge_color=edge_colors)
plt.show()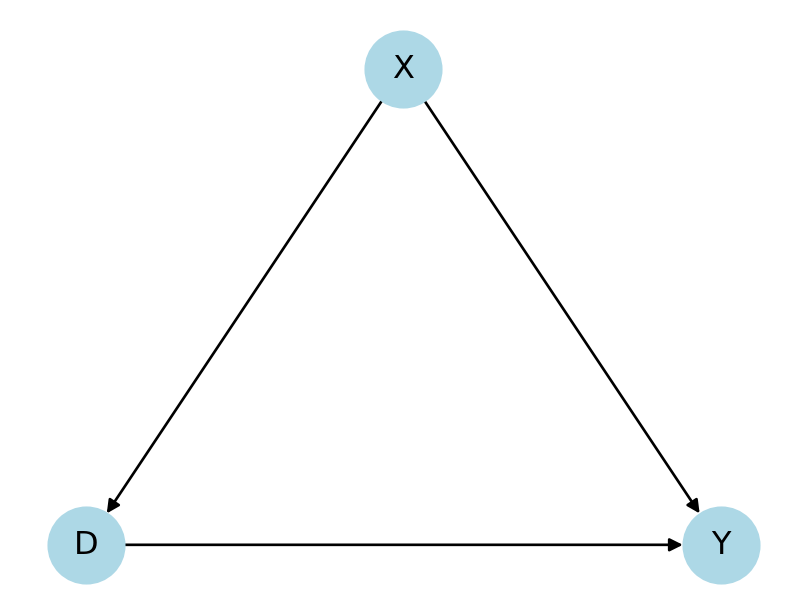
Motivation
What if the conditional independence assumption is violated?
Unobserved confounders U would introduce an omitted variable bias/ selection-into-treatment bias
Key questions:
- How strong would a confounding relationship need to be in order to change the conclusions of our analysis?
- Would such a confounding relationship be plausibly present in our data?
Code
import networkx as nx
import matplotlib.pyplot as plt
G = nx.DiGraph()
# Add nodes
G.add_node("D")
G.add_node("Y")
G.add_node("X")
G.add_node("U")
G.add_edge("D", "Y")
G.add_edge("X", "Y")
G.add_edge("X", "D")
G.add_edge("U", "D")
G.add_edge("U", "Y")
# Draw the graph
plt.figure(figsize=(4, 3))
pos = {"D": (0, 0), "Y": (2, 0), "X": (1,1), "U": (1,-1)}
edge_colors = ['black', 'black', 'black', 'red', 'blue']
nx.draw(G, pos, with_labels=True, node_size=800, node_color='lightblue',
edge_color=edge_colors)
plt.show()
Outlook: Sensitivity Analysis
- How strong would a confounding relationship need to be in order to change the conclusions of our analysis?
- Model the strength of the confounding relationships in terms of some sensitivity parameters, i.e.,
- \(U\) \(\rightarrow\) \(D\) and
- \(U\) \(\rightarrow\) \(Y\)
- Would such a confounding relationship be plausibly present in our data?
- Benchmarking framework
Code
import networkx as nx
import matplotlib.pyplot as plt
G = nx.DiGraph()
# Add nodes
G.add_node("D")
G.add_node("Y")
G.add_node("X")
G.add_node("U")
G.add_edge("D", "Y")
G.add_edge("X", "Y")
G.add_edge("X", "D")
G.add_edge("U", "D")
G.add_edge("U", "Y")
# Draw the graph
plt.figure(figsize=(4, 3))
pos = {"D": (0, 0), "Y": (2, 0), "X": (1,1), "U": (1,-1)}
edge_colors = ['black', 'black', 'black', 'red', 'blue']
nx.draw(G, pos, with_labels=True, node_size=800, node_color='lightblue',
edge_color=edge_colors)
plt.show()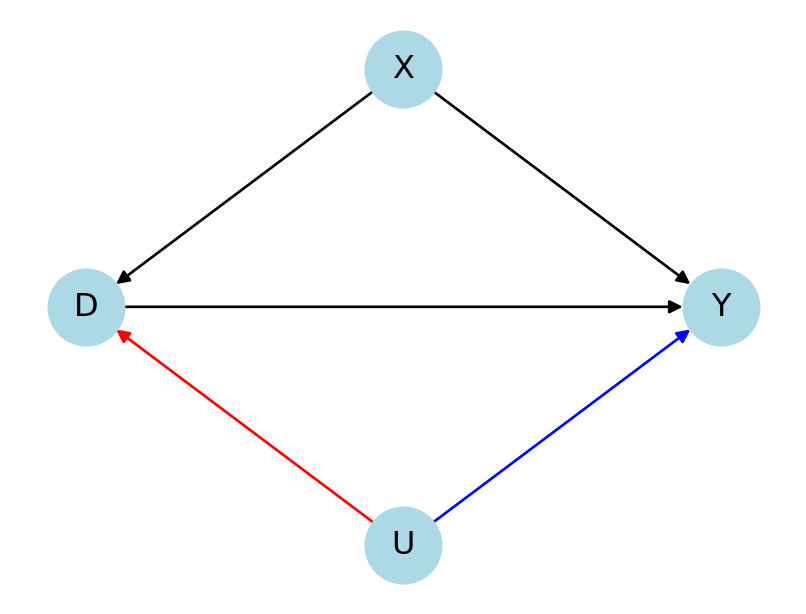
Example: Linear Regression Model
Consider a linear regression model with observed and unobserved confounders \(X\) and \(U\), respectively
\[ \begin{align*} Y &= \theta_0 D + \beta X + \gamma_1 U + \epsilon,\\ D &= \delta X + \gamma_2 U + \nu, \end{align*} \] with \(\epsilon\) and \(\nu\) being uncorrelated error terms and \(\theta_0\) corresponds to the average treatment effect of the (continuous) treatment \(D\) on \(Y\).
Short parameter \(\theta_{0,s}\) - corresponds to model using data \(\{Y, D, X\}\) (short model)1
Long parameter \(\theta_0\) - corresponds to model using data \(\{Y, D, X, U\}\) (long model)
\(\Rightarrow\) Omitted variable bias:
\[ \theta_{0,s} - \theta_{0} \]
Example: Linear Regression Model
\[ \begin{align*} Y &= \theta D + \beta X + \gamma_1 U + \epsilon,\\ D &= \delta X + \gamma_2 U + \nu, \end{align*} \]
- Simple idea for sensitivity analysis:
- Parametrically model / simulate the unobserved confounder \(U\) and derive sensitivity bounds according to different values of \(\gamma_1\) and \(\gamma_2\)
- ⚡ Problem: Sensitivity results will depend on the parametric model for \(U\), which might be too simplistic, for example,
- Binary or continuous \(U\)?
- Single or multiple confounders \(U\)?
- Correlation of \(U\) with \(X\)?
- Alternative: Use \(R^2\)-based sensitivity parameters (Cinelli and Hazlett 2020)
Example: Linear Regression Model
Sensitivity parameters in Cinelli and Hazlett (2020)
\(U\) \(\rightarrow\) \(D\): Share of residual variation of \(D\) explained by omitted confounder(s) \(U\), after taking \(X\) into account, \(R^2_{D\sim U|X}\)
\(U\) \(\rightarrow\) \(Y\): Share of residual variation of \(Y\) explained by \(U\), after taking \(X\) and \(D\) into account, \(R^2_{Y\sim U|D,X}\)
Code
import networkx as nx
import matplotlib.pyplot as plt
G = nx.DiGraph()
# Add nodes
G.add_node("D")
G.add_node("Y")
G.add_node("X")
G.add_node("U")
G.add_edge("D", "Y")
G.add_edge("X", "Y")
G.add_edge("X", "D")
G.add_edge("U", "D")
G.add_edge("U", "Y")
# Draw the graph
plt.figure(figsize=(4, 3))
pos = {"D": (0, 0), "Y": (2, 0), "X": (1,1), "U": (1,-1)}
edge_colors = ['black', 'black', 'black', 'red', 'blue']
nx.draw(G, pos, with_labels=True, node_size=800, node_color='lightblue',
edge_color=edge_colors)
plt.show()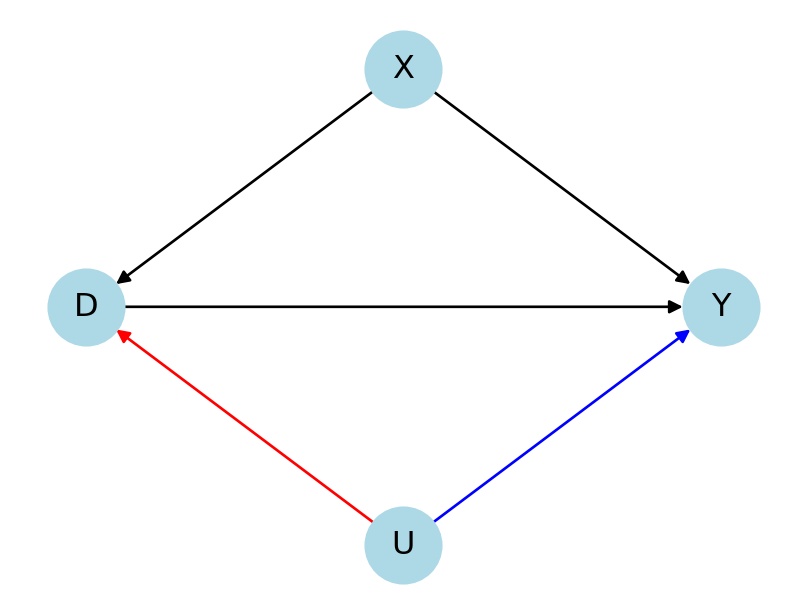
Example: Linear Regression Model
Results in Cinelli and Hazlett (2020)
Bounds for the omitted variable bias \(\theta_{0,s} - \theta_{0}\) and confidence intervals as based on values for these sensitivity parameters
Various measures to be reported:
- Robustness value
- Extreme scenarios
Visualization:
- Contour plots (Imbens 2003)
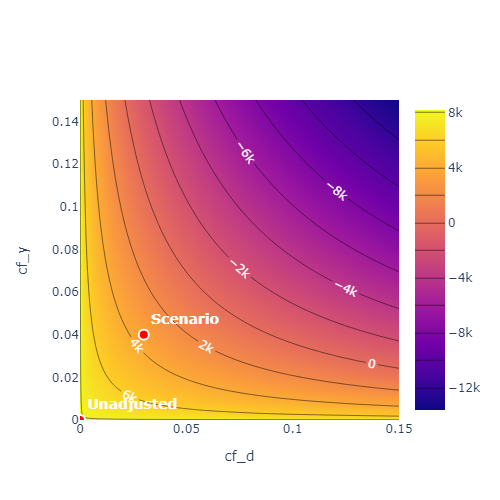
Example: Linear Regression Model
The approach by Cinelli and Hazlett (2020) can be used to derive sensitivity bounds depending on various specifications of the sensitivity parameters
However, how do we know if the values for these parameters are plausible?
Cinelli and Hazlett (2020) also develop a formal benchmarking framework to relate the values of the sensitivity parameters to observed confounders
Idea:
- Mimic omitting an important observed (benchmark) confounder and re-compute the values of the sensitivity parameters
- Use domain expertise to assess plausibility of the confounding scenarios
Example: Linear Regression Model
Benchmarking example
- Assume we know that \(X_1\) is a very important predictor for \(Y\) and \(D\), then we can calculate the benchmark values for the sensitivity parameters, i.e.,
\(X_1\) \(\rightarrow\) \(D\): Share of residual variation of \(D\) explained by the omitted confounder(s) \(X_1\), after taking \(X_{-1}\)into account,1 \(R^2_{D\sim X_1|X_{-1}}\)
\(X_1\) \(\rightarrow\) \(Y\): Share of residual variation of \(Y\) explained by \(X_1\), after taking \(X_{-1}\) and \(D\) into account, \(R^2_{Y\sim X_1|D,X_{-1}}\)
- Given the values for the benchmarking variable, we can judge whether critical values of the sensitivity parameters are plausible or not
Sensitivity Analysis for Causal ML
Sensitivity Analysis for Causal ML
The framework of Cinelli and Hazlett (2020) is very intuitive and powerful for the linear regression model
Here, linearity helps to endow the sensitivity parameters with an intuitive interpretation (partial \(R^2\) )
However, the framework itself does not directly expand to non-linear models, such as the interactive regression model
Sensitivity Analysis for Causal ML
Chernozhukov et al. (2022) propose a generalization of the sensitivity analysis framework to non-linear models and is suitable for ML-based estimation
We do not go into the formal details1 as the approach is technically evolved
We sketch the main ideas and demonstrate the implementation in DoubleML with an example
Sensitivity Analysis for Causal ML
The sensitivity parameters in Cinelli and Hazlett (2020) are formulated in terms of partial \(R^2\) measures which apply to linear relationships
However, we might want to model non-linear relationships
Example: Partially linear regression model
\[ \begin{align}\begin{aligned}Y = D \theta_0 + g_0(X) + \zeta, & &\mathbb{E}(\zeta | D,X) = 0,\\D = m_0(X) + V, & &\mathbb{E}(V | X) = 0,\end{aligned}\end{align} \] with nonlinear functions \(g_0\) and \(m_0\).
- Moreover, we would like to apply sensitivity analysis for
- Other causal models, such as the interactive regression model
- ML-based estimation
Sensitivity Analysis for Causal ML
Brief summary: Chernozhukov et al. (2022)
Generalization of the ideas in Cinelli and Hazlett (2020) to a broad class of causal models, including
- Partially linear regression
- Interactive regression model
- Difference-in-Differences
The approach is based on the so-called Riesz-Fréchet representation, which is related to the orthogonal score of a causal model (debiasing)
Sensitivity parameters are defined in terms of nonparametric partial \(R^2\)
Sensitivity Analysis for Causal ML
Brief summary: Chernozhukov et al. (2022)
Advantages
Various causal models (including non-separable models, like IRM)
ML-based estimation
Non-linear confounding relationships
Limitations
Technical complexity
Generalization comes at costs of interpretability (\(R^2\)?)
Sensitivity Analysis in DoubleML
Sensitivity Analysis in DoubleML
Let’s complete the 7th step of the DoubleML workflow example1
We obtained the following results for the ATE (IRM)
- Now, we wonder how robust these effects are with respect to unobserved confounding
Sensitivity Analysis in DoubleML
- At given values for the sensitivity parameters
cf_dandcf_y, we can compute bounds for- The parameter \(\theta_0\) and
- \((1-\alpha)\) confidence intervals
- The interpretation of the sensitivity parameters depends on the causal model
PLR
- \(U\) \(\rightarrow\) \(D\): Partial nonparametric \(R^2\) of \(U\) with \(D\), given \(X\)
- \(U\) \(\rightarrow\) \(Y\): Partial nonparametric \(R^2\) of \(U\) with \(Y\), given \(D\) and \(X\)
Code
import networkx as nx
import matplotlib.pyplot as plt
G = nx.DiGraph()
# Add nodes
G.add_node("D")
G.add_node("Y")
G.add_node("X")
G.add_node("U")
G.add_edge("D", "Y")
G.add_edge("X", "Y")
G.add_edge("X", "D")
G.add_edge("U", "D")
G.add_edge("U", "Y")
# Draw the graph
plt.figure(figsize=(4, 3))
pos = {"D": (0, 0), "Y": (2, 0), "X": (1,1), "U": (1,-1)}
edge_colors = ['black', 'black', 'black', 'red', 'blue']
nx.draw(G, pos, with_labels=True, node_size=800, node_color='lightblue',
edge_color=edge_colors)
plt.show()
Sensitivity Analysis in DoubleML
- At given values for the sensitivity parameters
cf_dandcf_y, we can compute bounds for- The parameter \(\theta_0\) and
- \((1-\alpha)\) confidence intervals
- The interpretation of the sensitivity parameters depends on the causal model
IRM
- \(U\) \(\rightarrow\) \(D\): Average gain in quality to predict \(D\) by using \(U\) in addition to \(X\) (relative)
- \(U\) \(\rightarrow\) \(Y\): Partial nonparametric \(R^2\) of \(U\) with \(Y\), given \(D\) and \(X\)
Code
import networkx as nx
import matplotlib.pyplot as plt
G = nx.DiGraph()
# Add nodes
G.add_node("D")
G.add_node("Y")
G.add_node("X")
G.add_node("U")
G.add_edge("D", "Y")
G.add_edge("X", "Y")
G.add_edge("X", "D")
G.add_edge("U", "D")
G.add_edge("U", "Y")
# Draw the graph
plt.figure(figsize=(4, 3))
pos = {"D": (0, 0), "Y": (2, 0), "X": (1,1), "U": (1,-1)}
edge_colors = ['black', 'black', 'black', 'red', 'blue']
nx.draw(G, pos, with_labels=True, node_size=800, node_color='lightblue',
edge_color=edge_colors)
plt.show()
Sensitivity Analysis in DoubleML
- Sensitivity analysis as of Chernozhukov et al. (2022) is implementented in the method
sensitivity_analysis()
================== Sensitivity Analysis ==================
------------------ Scenario ------------------
Significance Level: level=0.95
Sensitivity parameters: cf_y=0.03; cf_d=0.03, rho=1.0
------------------ Bounds with CI ------------------
CI lower theta lower theta theta upper CI upper
e401 2145.47168 4033.019801 8121.564764 12210.109726 14101.927365
------------------ Robustness Values ------------------
H_0 RV (%) RVa (%)
e401 0.0 5.870442 4.478475- Robustness value
RV: Minimum strength of the confounding relationship that would lead to an adjustment of the parameter bounds such that they include \(0\)1
- A confounding relationship with
cf_d\(=\)cf_y\(=5.87\%\) would suffice to set the lower bound for the ATE to \(0\)
Sensitivity Analysis in DoubleML
- Sensitivity analysis as of Chernozhukov et al. (2022) is implementented in the method
sensitivity_analysis()
================== Sensitivity Analysis ==================
------------------ Scenario ------------------
Significance Level: level=0.95
Sensitivity parameters: cf_y=0.03; cf_d=0.03, rho=1.0
------------------ Bounds with CI ------------------
CI lower theta lower theta theta upper CI upper
e401 2145.47168 4033.019801 8121.564764 12210.109726 14101.927365
------------------ Robustness Values ------------------
H_0 RV (%) RVa (%)
e401 0.0 5.870442 4.478475- Robustness value
RVa: Minimum strength of the confounding relationship that would lead to an adjustment of the (1-\(\alpha\)) confidence interval bounds such that they intersect with \(0\)
- A confounding relationship with
cf_d\(=\)cf_y\(=4.48\%\) would suffice to let the lower bound for the \(95\%\) confidence interval include \(0\) (=render effect non-significant)
Sensitivity Analysis in DoubleML
Visualization
Sensitivity Analysis in DoubleML
So far, we considered how we can perform sensitivity analysis for a given set of parameter values
However, we might wonder how we can choose plausible values for the sensitivity parameters
There are basically two ways to do this:
- Domain expertise: Use domain expertise to postulate certain values for the confounding scenarios
- Benchmarking: Mimic omitting an important observed (benchmark) confounder and re-compute the values of the sensitivity parameters
Sensitivity Analysis in DoubleML
Benchmarking
We consider the variable
incwhich measures individuals’ incomeFor more details, see Chapter 6 in Chernozhukov et al. (2022) as well as the notebook in the DoubleML Example Gallery
Sensitivity Analysis in DoubleML
Benchmarking
Sensitivity Analysis in DoubleML
Degree of adversity
When specifying the main confounding scenario in the
sensitivity_analysis()call, there is a parameter called \(\rho\) (degree of adversity)Informally speaking, \(\rho \in [-1, 1]\) measures the correlation between the deviations that are created by the confounder(s) in terms of the relationships
- \(U\) \(\rightarrow\) \(D\) and
- \(U\) \(\rightarrow\) \(Y\)
Intuitively, if the variations in \(D\) and \(Y\), which can be explained by the omitted variable \(U\), are uncorrelated, the resulting bias would be \(0\)
Sensitivity Analysis in DoubleML
Degree of adversity
\(\rho\) operates as a scaling factor in the omitted variable bias formula
Results are most conservative results with \(\rho = 1\) (default)
We can calibrate \(\rho\) during the empirical benchmarking procedure
Without further modification, the scenarios added to the contour plot are conservative (i.e., based on \(\rho = 1\))
Sensitivity Analysis in DoubleML
Benchmarking
- We can use the benchmarking scenario as the major confounding scenario and calibrate the contour plot according to \(\rho\)
================== Sensitivity Analysis ==================
------------------ Scenario ------------------
Significance Level: level=0.95
Sensitivity parameters: cf_y=0.14598551560922302; cf_d=0.17533317267482074, rho=0.21120271145715136
------------------ Bounds with CI ------------------
CI lower theta lower theta theta upper CI upper
e401 1206.761225 3127.196673 8121.564764 13115.932855 15041.493382
------------------ Robustness Values ------------------
H_0 RV (%) RVa (%)
e401 0.0 24.837451 19.469718Sensitivity Analysis in DoubleML
Benchmarking
- We can use the benchmarking scenario as the major confounding scenario and calibrate the contour plot according to \(\rho\)
Sensitivity Analysis in DoubleML
Conclusion: Workflow example
If we believe that we likely miss a confounder with similarly strong relationships with \(D\) and \(Y\) as the benchmark variable
inc, the results do not seem to be robustHowever, if we believe that excluding such a confounder is unlikely, we can be more confident in the results (e.g., compare to other benchmarking variables)
Sensitivity Analysis in DoubleML
General recommendations
We recommend to repeat the cross-fitting procedure several times (
n_rep) and to use multiple folds (n_folds)In the IRM, the propensity score predictions can render the sensitivity analysis results unstable, so propensity score trimming might be helpful
The benchmarking procedure requires re-estimation of the underlying models and, hence, can become quite expensive under computational considerations
General comment 💡
- Conclusions from sensitivity analyses will generally not be unambiguous - they depend on the context of the study and need to be interpreted based on domain expertise
Appendix
Appendix: Sensitivity Analysis for Causal ML
- Nonparametric partial \(R^2\)
\[ \frac{\textrm{Var}(\mathbb{E}[Y|D,X,A]) - \textrm{Var}(\mathbb{E}[Y|D,X])}{\textrm{Var}(Y)-\textrm{Var}(\mathbb{E}[Y|D,X])} \]
Appendix: Sensitivity Analysis for IRM
Riesz-Fréchet representation for IRM (ATE)
\[ \begin{align} \theta_0 &= \mathbb{E} (m(W, g))\\ &= \mathbb{E} (g(1, X) - g(0, X)), \end{align} \] with \(g(d,X)=\mathbb{E}[Y|D=d, X]\). The Riesz-representation theorem says that we can re-write the \(\theta_0\) as \[ \theta_0 = \mathbb{E}[g_0(W)\underbrace{\alpha_0(W)}_{RR}]. \] In the IRM we have \[ \alpha_0(W) = \frac{D}{m(X)} - \frac{1-D}{1-m(X)}. \]
Appendix: Sensitivity Analysis for IRM
Riesz-Frechet representation for IRM (ATE)
The Riesz-Representer (RR) points down a debiased / orthogonal score function, see Chernozhukov, Newey, and Singh (2022).1
\[ \begin{align} \psi(W, \theta_0, g, \alpha) = & m(W, g ) - \theta_0 + \alpha(W) \{ Y - g(X)\} \end{align} \] For the ATE in the IRM (Example 3 in Chernozhukov, Newey, and Singh (2022)), we have
\[ \begin{align}\begin{aligned}\psi(\cdot) := & g(1,X) - g(0,X) - \theta \\ & + \frac{D (Y - g(1,X))}{m(X)} - \frac{(1 - D)(Y - g(0,X))}{1 - m(x)} \end{aligned}\end{align} \]
which is the doubly-robust score.
Appendix: Sensitivity Analysis for IRM
- Sensitivity parameter
cf_d\(:= \frac{C_D^2}{1+C_D^2}\) with
\[ C_D^2= \frac{\mathbb{E}\Big[\big(P(D=1|X,A)(1-P(D=1|X,A))\big)^{-1}\Big]}{\mathbb{E}\Big[\big(P(D=1|X)(1-P(D=1|X))\big)^{-1}\Big]} - 1 \]
References
References
DoubleML - Tools for Causality 2023